Abstract
Introduction: The confirmation of a diagnosis of myelodysplastic syndrome (MDS) can be very difficult based on morphology alone. The detection of cytogenetic abnormalities or the presence of mutations in one or more of the myeloid-related genes can provide significant help in confirming this diagnosis. However, some patients with MDS may lack mutations or cytogenetic abnormalities. Cell free DNA (cfDNA) and microRNA are used extensively in testing for molecular abnormalities in hematologic and solid tumors, but little is known about the value of cell free mRNA (cfRNA) in liquid biopsy. We explored the use of targeted enrichment and Next Generation Sequencing (NGS) of cfRNA in the RNA profiling and diagnosis of MDS.
Methods: We used NucliSenS EasyMAG (bioMérieux, Marcy-l'Étoile, France) automated platform for extracting total nucleic acid from peripheral blood plasma collected in EDTA. We used the TruSight RNA Pan-Cancer Panel (Illumina, San Diego, CA) for detecting fusion, expression, and mutations in 1385 genes. The Pan-Cancer Panel is a hybridization based targeted panel, therefore alignments and gene expression are calculated only to the 1385 genes as per manufacturer recommendations. Each sample was deeply sequenced (>55M reads) using Illumina paired-end 75 bp sequencing and duplicate reads were removed before fragment per kilobase of transcript per million mapped reads (FPKMs) were calculated for each gene. Samples from 23 patients were analyzed including 19 patients confirmed by molecular, cytogenetic, and morphologic methods to have MDS or chronic myelomonocytic leukemia (CMML) and 4 individuals who had one or more cytopenia, but MDS was ruled out by molecular mutation analysis, cytogenetics, as well as morphology. Expression levels were first normalized using ABL1 and PAX5 (i.e., regressing each gene on ABL1 and PAX5 jointly and using the residuals instead of the raw data) and models for distinguishing MDS from non-MDS were developed.
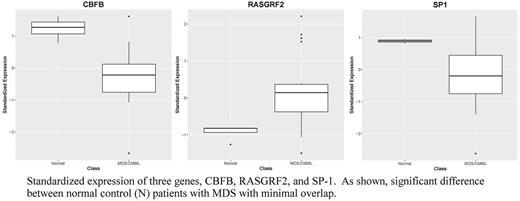
Results: In this study, only expression levels were considered and not the mutational profile. We used t-tests and examined the statistical significance of each gene separately. Then we selected genes with P-values < 0.05 to filter out non-significant genes, resulting in 49 significant genes. After adjusting for multiple hypothesis testing, only three genes remained significant with false discovery rate set at 0.1. The three genes were: CBFB, RASGRF2, and SP1 . Using a logistic regression model based on these three variables, we were able to obtain accuracy rate of 87.0% and AUC (area under the ROC curve) of 92.1% based on the leave-one-out testing. This model identified all the normal cases correctly, but misclassified 3 MDS cases as normal. Alternatively, using the top 3 principal components based on the selected 49 significant genes in a linear regression model, we can improve prediction and AUC to 95.6% and 96.7%, respectively, based on the leave-one-out testing. In this model, only one MDS case (out of 19) was misclassified as normal, while all the normal cases were classified correctly.
Conclusion: Although additional studies and validation are needed, RNA profiling using targeted enrichment NGS of cfRNA is potentially useful in the classification and diagnosis of MDS. This model can be expanded to incorporate an entire mutational profile assessment. This approach has the potential to be used not only for MDS diagnosis, but also in the diagnosis, prognostic prediction and response to therapy of a variety of myeloid and lymphoid neoplasms.
Wong: NeoGenomics: Employment. Funari: NeoGenomics: Employment. Ma: NeoGenomics: Employment. De Dios: NeoGenomics: Employment. Blocker: NeoGenomics: Employment. Albitar: NeoGenomics: Employment.
Author notes
Asterisk with author names denotes non-ASH members.